4. Firedrake 中的并行计算#
4.1. 终端中并行执行 Firedrake 程序#
Firedrake 启动并行计算只需在终端中运行以下命令:
mpiexec -n <number-of-process> python3 /path/to/your/script.py
下面以求解 Poisson 方程的程序为例, 将以下代码保存为文件 poisson.py
from firedrake import *
N = 4
test_mesh = RectangleMesh(nx=N, ny=N, Lx=1, Ly=1)
x, y = SpatialCoordinate(test_mesh)
f = sin(pi*x)*sin(pi*y)
g = Constant(0)
V = FunctionSpace(test_mesh, 'CG', degree=1)
u, v = TrialFunction(V), TestFunction(V)
a = inner(grad(u), grad(v))*dx
L = inner(f, v)*dx
bc = DirichletBC(V, g=g, sub_domain='on_boundary')
u_h = Function(V, name='u_h')
solve(a == L, u_h, bcs=bc)
output = VTKFile('data/result.pvd')
output.write(u_h)
若使用 2 个进程进行计算, 在激活的 Firedrake 环境中运行:
mpiexec -n 2 python3 poisson.py
求解结果将保存在 data/result.pvd 文件中. 在 data 目录下会生成一个 result 文件夹,
计算结果保存在该文件夹中, 每个进程对应一个结果文件. result.pvd 文件只是这些结果文件的索引.
Firedrake 使用 PETSc 的 DMPlex 来管理网格. 在并行计算中, 计算区域会被划分为多个子区域 (默认情况下区域间有交叠), 并分配给不同的进程, 因此每个进程的结果文件仅包含该进程对应区域的结果.
4.1.1. 并行时的一些注意事项#
4.1.1.1. 并行时的输出#
第一个进程输出 (其他进程的调用会被忽略)
PETSc.Sys.Print("This is the message will only show once!")
多个进程同步输出
rank, size = COMM_WORLD.rank, COMM_WORLD.size PETSc.Sys.syncPrint(f"[{rank}/{size}] This is the message from rank {rank}!") if rank == 0: PETSc.Sys.syncPrint(f"[{rank}/{size}] Message from rank {rank}!") PETSc.Sys.syncFlush()
4.1.1.2. 仅在指定进程上运算#
如果需要在某个特定进程上执行某些操作或计算, 可以使用条件语句. 例如, 只在第 0 号进程上进行绘图操作:
if COMM_WORLD.rank == 0:
triplot(...)
4.2. 在 Jupyter-notebook/Jupyter-lab 中并行执行 Firedrake 代码#
在 Jupyter 交互环境中, 我们可以方便地对串行代码进行测试, 从而进行快速验证.
为了在 Jupyter 环境下运行并行程序, ipyparallel 包提供了强大的支持,
它能够帮助我们在 Jupyter 中验证并行代码, 同时加深我们对 Firedrake 并行机制的理解.
4.2.1. 测试 ipyparallel#
启动并连接 Cluster.
import ipyparallel as ipp
cluster = ipp.Cluster(engines="mpi", n=2)
client = cluster.start_and_connect_sync()
Starting 2 engines with <class 'ipyparallel.cluster.launcher.MPIEngineSetLauncher'>
在配置好 ipyparallel 并启动集群后, 可以通过 Jupyter 中的魔法指令 %%px 在多个进程上并行执行代码.
%%px 是 ipyparallel 提供的魔法命令, 能够在所有并行引擎上同时执行代码.
--block 参数用于同步执行代码, 确保代码在所有进程上完成后再继续执行后续命令.
%%px --block
from firedrake import *
from firedrake.petsc import PETSc
from mpi4py import MPI
N = 4
mesh = RectangleMesh(N, N, 1, 1)
rank, size = mesh.comm.rank, mesh.comm.size
PETSc.Sys.syncPrint(f"[{rank}/{size}] This is the message from rank {rank}!")
PETSc.Sys.syncFlush()
PETSc.Sys.Print(f"[{rank}/{size}] This message only from rank 0!")
[stdout:0] [0/2] This is the message from rank 0!
[1/2] This is the message from rank 1!
[0/2] This message only from rank 0!
4.3. 使用 ipyparallel 观察串行和并行过程#
4.3.1. 生成网格并画出网格#
4.3.1.1. 串行#
from firedrake import *
from firedrake.pyplot import triplot
from firedrake.petsc import PETSc
from mpi4py import MPI
import matplotlib.pyplot as plt
N = 4
mesh = RectangleMesh(N, N, 1, 1)
mesh.topology_dm.view()
fig, axes = plt.subplots(figsize=[4, 3])
c = triplot(mesh, axes=axes)
xlim = axes.set_xlim([-0.1,1.1])
ylim = axes.set_ylim([-0.1,1.1])
DM Object: firedrake_default_topology 1 MPI process
type: plex
firedrake_default_topology in 2 dimensions:
Number of 0-cells per rank: 25
Number of 1-cells per rank: 56
Number of 2-cells per rank: 32
Labels:
celltype: 3 strata with value/size (0 (25), 1 (56), 3 (32))
depth: 3 strata with value/size (0 (25), 1 (56), 2 (32))
Face Sets: 4 strata with value/size (1 (9), 2 (9), 3 (9), 4 (9))
exterior_facets: 1 strata with value/size (1 (32))
interior_facets: 1 strata with value/size (1 (63))
pyop2_core: 1 strata with value/size (1 (113))
pyop2_owned: 0 strata with value/size ()
pyop2_ghost: 0 strata with value/size ()

4.3.1.2. 并行#
%%px --block
from firedrake import *
from firedrake.pyplot import triplot
from firedrake.petsc import PETSc
from mpi4py import MPI
import matplotlib.pyplot as plt
N = 4
mesh = RectangleMesh(N, N, 1, 1)
mesh.topology_dm.view()
fig, axes = plt.subplots(figsize=[4, 3])
c = triplot(mesh, axes=axes)
xlim = axes.set_xlim([-0.1,1.1])
ylim = axes.set_ylim([-0.1,1.1])
[output:1]
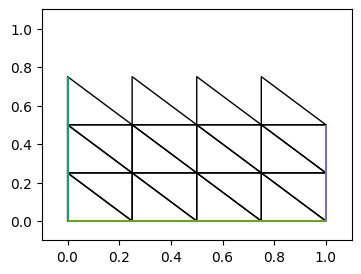
[stdout:0] DM Object: firedrake_default_topology 2 MPI processes
type: plex
firedrake_default_topology in 2 dimensions:
Number of 0-cells per rank: 19 19
Number of 1-cells per rank: 38 38
Number of 2-cells per rank: 20 20
Labels:
depth: 3 strata with value/size (0 (19), 1 (38), 2 (20))
celltype: 3 strata with value/size (0 (19), 1 (38), 3 (20))
Face Sets: 3 strata with value/size (1 (5), 2 (7), 4 (9))
exterior_facets: 1 strata with value/size (1 (19))
interior_facets: 1 strata with value/size (1 (47))
pyop2_core: 1 strata with value/size (1 (26))
pyop2_owned: 1 strata with value/size (1 (26))
pyop2_ghost: 1 strata with value/size (1 (25))
[output:0]
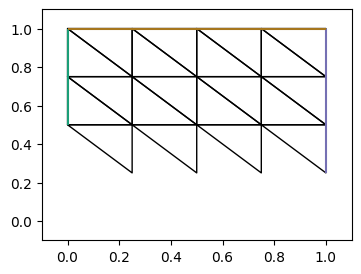
可以看到上面并行中两个网格图是整体网格的一部分, 且有重叠部分.
4.3.2. 定义变分问题#
4.3.2.1. 串行#
V1 = VectorFunctionSpace(mesh, 'CG', 1)
V2 = FunctionSpace(mesh, 'CG', 2)
W = MixedFunctionSpace([V1, V2]) # W = V1*V2
u1, u2 = TrialFunctions(W)
v1, v2 = TestFunctions(W)
a = dot(u1, v1)*dx + u2*v2*dx
x, y = SpatialCoordinate(mesh)
f = dot(as_vector((sin(x), cos(y))), v1)*dx + cos(y)*v2*dx
bc = DirichletBC(W.sub(0), 0, 1)
uh = Function(W)
problem = LinearVariationalProblem(a, f, uh, bcs=bc)
4.3.2.2. 并行#
%%px --block
V1 = VectorFunctionSpace(mesh, 'CG', 1)
V2 = FunctionSpace(mesh, 'CG', 2)
W = MixedFunctionSpace([V1, V2]) # W = V1*V2
u1, u2 = TrialFunctions(W)
v1, v2 = TestFunctions(W)
a = dot(u1, v1)*dx + u2*v2*dx
x, y = SpatialCoordinate(mesh)
f = dot(as_vector((sin(x), cos(y))), v1)*dx + cos(y)*v2*dx
bc = DirichletBC(W.sub(0), 0, 1)
uh = Function(W)
problem = LinearVariationalProblem(a, f, uh, bcs=bc)
4.3.3. 函数空间维度#
函数空间 V1 和 V2 中的自由度, 可以通过调用函数 dim 得到
rank, size = mesh.comm.rank, mesh.comm.size
PETSc.Sys.Print(f'Number of dofs of V1: {V1.dim()}')
PETSc.Sys.syncPrint(f'[{rank}/{size}] V1 Node count: {V1.node_count}; V1 Dof count: {V1.dof_count}')
PETSc.Sys.syncFlush()
Number of dofs of V1: 50
[0/1] V1 Node count: 25; V1 Dof count: 50
%%px --block
rank, size = mesh.comm.rank, mesh.comm.size
PETSc.Sys.Print(f'Number of dofs of V1: {V1.dim()}')
PETSc.Sys.syncPrint(f'[{rank}/{size}] V1 Node count: {V1.node_count}; V1 Dof count: {V1.dof_count}; V1 dim: {V1.dim()}')
PETSc.Sys.syncFlush()
[stdout:0] Number of dofs of V1: 50
[0/2] V1 Node count: 19; V1 Dof count: 38; V1 dim: 50
[1/2] V1 Node count: 19; V1 Dof count: 38; V1 dim: 50
%%px --block
f_v1 = Function(V1)
with f_v1.dat.vec_ro as vec:
PETSc.Sys.syncPrint(f'[{rank}/{size}] {vec.getSizes()}')
PETSc.Sys.syncFlush()
[stdout:0] [0/2] (20, 50)
[1/2] (30, 50)
%%px --block
f_v1.dat.data.shape, f_v1.dat.data_with_halos.shape
Out[0:6]: ((10, 2), (19, 2))
Out[1:6]: ((15, 2), (19, 2))
4.3.4. 节点集和自由度数据集#
PETSc.Sys.syncPrint(f'[{rank}/{size}] V1: {str(V1.node_set)}')
PETSc.Sys.syncPrint(f'[{rank}/{size}] {repr(V1.node_set)}')
PETSc.Sys.syncPrint(f'[{rank}/{size}] V1: {str(V1.dof_dset)}')
PETSc.Sys.syncPrint(f'[{rank}/{size}] {repr(V1.dof_dset)}')
PETSc.Sys.syncFlush()
[0/1] V1: OP2 Set: set_#x7fa3fcc9c890 with size 25
[0/1] Set((np.int64(25), np.int64(25), np.int64(25)), 'set_#x7fa3fcc9c890')
[0/1] V1: OP2 DataSet: None_nodes_dset on set OP2 Set: set_#x7fa3fcc9c890 with size 25, with dim (2,), False
[0/1] DataSet(Set((np.int64(25), np.int64(25), np.int64(25)), 'set_#x7fa3fcc9c890'), (2,), 'None_nodes_dset', False)
%%px --block
PETSc.Sys.syncPrint(f'[{rank}/{size}] V1: {str(V1.node_set)}')
PETSc.Sys.syncPrint(f'[{rank}/{size}] {repr(V1.node_set)}')
PETSc.Sys.syncPrint(f'[{rank}/{size}] V1: {str(V1.dof_dset)}')
PETSc.Sys.syncPrint(f'[{rank}/{size}] {repr(V1.dof_dset)}')
PETSc.Sys.syncFlush()
[stdout:0] [0/2] V1: OP2 Set: set_#x7f86527cec30 with size 10
[0/2] Set((np.int64(5), np.int64(10), np.int64(19)), 'set_#x7f86527cec30')
[0/2] V1: OP2 DataSet: None_nodes_dset on set OP2 Set: set_#x7f86527cec30 with size 10, with dim (2,), False
[0/2] DataSet(Set((np.int64(5), np.int64(10), np.int64(19)), 'set_#x7f86527cec30'), (2,), 'None_nodes_dset', False)
[1/2] V1: OP2 Set: set_#x7fbf544618b0 with size 15
[1/2] Set((np.int64(10), np.int64(15), np.int64(19)), 'set_#x7fbf544618b0')
[1/2] V1: OP2 DataSet: None_nodes_dset on set OP2 Set: set_#x7fbf544618b0 with size 15, with dim (2,), False
[1/2] DataSet(Set((np.int64(10), np.int64(15), np.int64(19)), 'set_#x7fbf544618b0'), (2,), 'None_nodes_dset', False)
4.3.4.1. Size of Set and Data Set#
# Reference:
# https://github.com/OP2/PyOP2/blob/31471a606a852aed250b05574d1fc2a2874eec31/pyop2/types/set.py#L30
#
# The division of set elements is:
#
# [0, CORE)
# [CORE, OWNED)
# [OWNED, GHOST)
#
# Attribute of dof_dset
# core_size: Core set size. Owned elements not touching halo elements.
# size: Set size, owned elements.
# total_size: Set size including ghost elements.
# sizes: (core_size, size, total_size)
node_set = V1.node_set
msg = f'core size: {node_set.core_size}, size: {node_set.size}, total size: {node_set.total_size}'
# another size: node_set.constrained_size
PETSc.Sys.syncPrint(f'[{rank}/{size}] {msg}')
PETSc.Sys.syncFlush()
[0/1] core size: 25, size: 25, total size: 25
%%px --block
node_set = V1.node_set
msg = f'core size: {node_set.core_size}, size: {node_set.size}, total size: {node_set.total_size}'
PETSc.Sys.syncPrint(f'[{rank}/{size}] {msg}')
PETSc.Sys.syncFlush()
[stdout:0] [0/2] core size: 5, size: 10, total size: 19
[1/2] core size: 10, size: 15, total size: 19
dof_dset = V1.dof_dset
size_msg = f'core size: {dof_dset.core_size}, size: {dof_dset.size}, total size: {dof_dset.total_size}'
# dim: shape tuple of the values for each element, cdim: product of dim tuple
dim_msg = f'dim: {dof_dset.dim}, cdim: {dof_dset.cdim}'
PETSc.Sys.syncPrint(f'[{rank}/{size}] {size_msg}, {dim_msg}')
PETSc.Sys.syncFlush()
[0/1] core size: 25, size: 25, total size: 25, dim: (2,), cdim: 2
%%px --block
dof_dset = V1.dof_dset
size_msg = f'core size: {dof_dset.core_size}, size: {dof_dset.size}, total size: {dof_dset.total_size}'
# dim: shape tuple of the values for each element, cdim: product of dim tuple
dim_msg = f'dim: {dof_dset.dim}, cdim: {dof_dset.cdim}'
PETSc.Sys.syncPrint(f'[{rank}/{size}] {size_msg}, {dim_msg}')
PETSc.Sys.syncFlush()
[stdout:0] [0/2] core size: 5, size: 10, total size: 19, dim: (2,), cdim: 2
[1/2] core size: 10, size: 15, total size: 19, dim: (2,), cdim: 2
4.3.4.2. ISES of Data Set#
# field_ises:
# https://github.com/OP2/PyOP2/blob/31471a606a852aed250b05574d1fc2a2874eec31/pyop2/types/dataset.py#L145
# A list of PETSc ISes defining the global indices for each set in the DataSet.
# Used when extracting blocks from matrices for solvers.
#
# local_ises:
# A list of PETSc ISes defining the local indices for each set in the DataSet.
# Used when extracting blocks from matrices for assembly.
#
local_ises_msg = f'{[_.getIndices() for _ in W.dof_dset.local_ises]}'
PETSc.Sys.syncPrint(f'[{rank}/{size}] {local_ises_msg}')
PETSc.Sys.syncFlush()
[0/1] [array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49],
dtype=int32), array([ 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62,
63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75,
76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101,
102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114,
115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127,
128, 129, 130], dtype=int32)]
%%px --block
local_ises_msg = f'{[_.getIndices() for _ in W.dof_dset.local_ises]}'
PETSc.Sys.syncPrint(f'[{rank}/{size}] {local_ises_msg}')
PETSc.Sys.syncFlush()
[stdout:0] [0/2] [array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37], dtype=int32), array([38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54,
55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71,
72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88,
89, 90, 91, 92, 93, 94], dtype=int32)]
[1/2] [array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37], dtype=int32), array([38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54,
55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71,
72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88,
89, 90, 91, 92, 93, 94], dtype=int32)]
field_ises_msg = f'{[_.getIndices() for _ in W.dof_dset.field_ises]}'
PETSc.Sys.syncPrint(f'[{rank}/{size}] {field_ises_msg}')
PETSc.Sys.syncFlush()
[0/1] [array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49],
dtype=int32), array([ 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62,
63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75,
76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101,
102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114,
115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127,
128, 129, 130], dtype=int32)]
%%px --block
field_ises_msg = f'{[_.getIndices() for _ in W.dof_dset.field_ises]}'
PETSc.Sys.syncPrint(f'[{rank}/{size}] {field_ises_msg}')
PETSc.Sys.syncFlush()
[stdout:0] [0/2] [array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19], dtype=int32), array([20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53,
54, 55], dtype=int32)]
[1/2] [array([56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72,
73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85], dtype=int32), array([ 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98,
99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111,
112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124,
125, 126, 127, 128, 129, 130], dtype=int32)]
4.3.4.3. Local to Global Map#
# https://gitlab.com/petsc/petsc/-/blob/release/src/binding/petsc4py/src/petsc4py/PETSc/DM.pyx#L1777
# setSection = setLocalSection
# getSection = getLocalSection
# setDefaultSection = setLocalSection
# getDefaultSection = getLocalSection
halo = V1.dof_dset.halo
# sf = halo.dm.getPointSF()
# sf.view()
halo.local_to_global_numbering
# halo.dm.getLocalSection().view()
# halo.dm.getGlobalSection().view()
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24], dtype=int32)
%%px --block
halo = V1.dof_dset.halo
# sf = halo.dm.getPointSF()
# sf.view()
halo.local_to_global_numbering
# halo.dm.getLocalSection().view()
# halo.dm.getGlobalSection().view()
Out[1:12]:
array([10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 9, 8,
7, 6], dtype=int32)
Out[0:12]:
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 24, 23, 19, 22, 17, 21, 15,
20, 13], dtype=int32)
# lgmap:
# https://github.com/OP2/PyOP2/blob/31471a606a852aed250b05574d1fc2a2874eec31/pyop2/types/dataset.py#L111
# A PETSc LGMap mapping process-local indices to global indices
[V1.dof_dset.lgmap.apply(_) for _ in V1.dof_dset.local_ises]
[array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49],
dtype=int32)]
%%px --block
# [W.dof_dset.lgmap.apply(_) for _ in W.dof_dset.local_ises]
[V1.dof_dset.lgmap.apply(_) for _ in V1.dof_dset.local_ises]
Out[0:13]:
[array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 48, 49, 46, 47, 38, 39, 44, 45, 34, 35, 42, 43, 30, 31,
40, 41, 26, 27], dtype=int32)]
Out[1:13]:
[array([20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 18, 19, 16, 17,
14, 15, 12, 13], dtype=int32)]
V1.dof_dset.lgmap.indices
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49],
dtype=int32)
4.3.4.4. Layout Vector of Data Set#
# dof_dset.layout_vec.getSizes()
vec_msg = f'Local Size: {dof_dset.layout_vec.getLocalSize()}, Size: {dof_dset.layout_vec.getSize()}'
PETSc.Sys.syncPrint(f'[{rank}/{size}] {vec_msg}')
PETSc.Sys.syncFlush()
[0/1] Local Size: 50, Size: 50
%%px --block
# dof_dset.layout_vec.getSizes()
vec_msg = f'Local Size: {dof_dset.layout_vec.getLocalSize()}, Size: {dof_dset.layout_vec.getSize()}'
PETSc.Sys.syncPrint(f'[{rank}/{size}] {vec_msg}')
PETSc.Sys.syncFlush()
[stdout:0] [0/2] Local Size: 20, Size: 50
[1/2] Local Size: 30, Size: 50
4.3.5. 定义 Solver#
Firedrake 使用 PETSc 的 SNES 对非线性问题 (包括线性问题)
进行求解,并提供了4个回调函数用于在组装前后修改 Jacobian (\(J = F'(x)\)) 和残差 F,这4个函数分别为
pre_jacobian_callback,post_jacobian_callback, pre_function_callback, post_function_callback.
def post_jacobian_callback(X, J, appctx=None):
# appctx: user data
# X: vector (gauss value)
# J: Jacobian, i.e., F'(x)
#
# mat reference:
# https://petsc.org/main/petsc4py/reference/petsc4py.PETSc.Mat.html
PETSc.Sys.Print("post jacobian callback begin")
rank, size = J.comm.rank, J.comm.size
PETSc.Sys.syncPrint(f" [{rank}/{size}] appctx: {appctx}")
PETSc.Sys.syncFlush()
# J.setValueLocal(i, j, 1, PETSc.InsertMode.ADD_VALUES)
# J.assemble()
PETSc.Sys.Print("post jacobian callback end")
def post_function_callback(X, F, appctx=None):
# appctx: user data
# X: vector (gauss value)
# F: residual, i.e., F(x)
#
# vec reference:
# https://petsc.org/main/petsc4py/reference/petsc4py.PETSc.Vec.html
PETSc.Sys.Print("post function callback begin")
PETSc.Sys.syncPrint(f" [{rank}/{size}] appctx: {appctx}")
PETSc.Sys.syncFlush()
PETSc.Sys.Print("post function callback end")
appctx = {
'mesh': mesh,
'msg': 'The msg from appctx'
}
solver = LinearVariationalSolver(problem,
post_jacobian_callback=partial(post_jacobian_callback, appctx=appctx),
post_function_callback=partial(post_function_callback, appctx=appctx))
solver.solve()
post function callback begin
[0/1] appctx: {'mesh': Mesh(VectorElement(FiniteElement('Lagrange', triangle, 1), dim=2), 0), 'msg': 'The msg from appctx'}
post function callback end
post jacobian callback begin
[0/1] appctx: {'mesh': Mesh(VectorElement(FiniteElement('Lagrange', triangle, 1), dim=2), 0), 'msg': 'The msg from appctx'}
post jacobian callback end
%%px --block
def post_jacobian_callback(X, J, appctx=None):
# appctx: user data
# X: vector (gauss value)
# J: mat
#
# mat reference:
# https://petsc.org/main/petsc4py/reference/petsc4py.PETSc.Mat.html
PETSc.Sys.Print("post jacobian callback begin")
rank, size = J.comm.rank, J.comm.size
PETSc.Sys.syncPrint(f" [{rank}/{size}] appctx: {appctx}")
PETSc.Sys.syncFlush()
# J.setValueLocal(i, j, 1, PETSc.InsertMode.ADD_VALUES)
# J.assemble()
PETSc.Sys.Print("post jacobian callback end")
def post_function_callback(X, F, appctx=None):
# appctx: user data
# X: vector (gauss value)
# F: vector
#
# vec reference:
# https://petsc.org/main/petsc4py/reference/petsc4py.PETSc.Vec.html
PETSc.Sys.Print("post function callback begin")
PETSc.Sys.syncPrint(f" [{rank}/{size}] appctx: {appctx}")
PETSc.Sys.syncFlush()
PETSc.Sys.Print("post function callback end")
appctx = {
'mesh': mesh,
'msg': 'The msg from appctx'
}
solver = LinearVariationalSolver(problem,
post_jacobian_callback=partial(post_jacobian_callback, appctx=appctx),
post_function_callback=partial(post_function_callback, appctx=appctx))
solver.solve()
[stdout:0] post function callback begin
[0/2] appctx: {'mesh': Mesh(VectorElement(FiniteElement('Lagrange', triangle, 1), dim=2), 2), 'msg': 'The msg from appctx'}
[1/2] appctx: {'mesh': Mesh(VectorElement(FiniteElement('Lagrange', triangle, 1), dim=2), 2), 'msg': 'The msg from appctx'}
post function callback end
post jacobian callback begin
[0/2] appctx: {'mesh': Mesh(VectorElement(FiniteElement('Lagrange', triangle, 1), dim=2), 2), 'msg': 'The msg from appctx'}
[1/2] appctx: {'mesh': Mesh(VectorElement(FiniteElement('Lagrange', triangle, 1), dim=2), 2), 'msg': 'The msg from appctx'}
post jacobian callback end
4.3.5.1. Context of the Solver#
ctx = solver._ctx
PETSc.Sys.syncPrint(f'[{rank}/{size}] Assembler: {ctx._assembler_jac}')
PETSc.Sys.syncPrint(f'[{rank}/{size}] Matrix Size: {ctx._jac.petscmat.getSizes()}')
PETSc.Sys.syncFlush()
[0/1] Assembler: <firedrake.assemble.ExplicitMatrixAssembler object at 0x7fa3948f3440>
[0/1] Matrix Size: ((131, 131), (131, 131))
%%px --block
ctx = solver._ctx
PETSc.Sys.syncPrint(f'[{rank}/{size}] Assembler: {ctx._assembler_jac}')
PETSc.Sys.syncPrint(f'[{rank}/{size}] Matrix Size: {ctx._jac.petscmat.getSizes()}')
PETSc.Sys.syncFlush()
[stdout:0] [0/2] Assembler: <firedrake.assemble.ExplicitMatrixAssembler object at 0x7f8694e07ad0>
[0/2] Matrix Size: ((56, 131), (56, 131))
[1/2] Assembler: <firedrake.assemble.ExplicitMatrixAssembler object at 0x7fbf0ddceb10>
[1/2] Matrix Size: ((75, 131), (75, 131))
4.3.5.2. Matrix 组装#
矩阵存储分配相关函数
Firedrake method: ExplicitMatrixAssembler.allocate
PyOp2 class: Sparsity
PyOp2 function: build_sparsity
PyOp2 class: Mat
更多组装细节请看 矩阵组装内核