9. DMPlex 和 Mesh#
准备并行环境
import ipyparallel as ipp
cluster = ipp.Cluster(profile="mpi", n=2)
client = cluster.start_and_connect_sync()
Starting 2 engines with <class 'ipyparallel.cluster.launcher.LocalEngineSetLauncher'>
%%px --block
from firedrake import *
from firedrake.petsc import PETSc
from mpi4py import MPI
在串行环境中导入必要的包
from firedrake import *
from firedrake.petsc import PETSc
from mpi4py import MPI
9.1. 创建 DMPlex#
9.1.1. 底层创建方法#
plex = PETSc.DMPlex().create()
plex.setDimension(2)
plex.setChart(0, 11)
# plex.setConeSize(point, number of points that cover the point)
plex.setConeSize(0, 3)
plex.setConeSize(1, 3)
plex.setConeSize(6, 2)
plex.setConeSize(7, 2)
plex.setConeSize(8, 2)
plex.setConeSize(9, 2)
plex.setConeSize(10, 2)
plex = plex.setUp() # plex.setUp() return self
# plex.setCone(point, [points that cover the point])
plex.setCone(0, [6, 7, 8])
plex.setCone(1, [7, 9, 10])
plex.setCone(6, [2, 3])
plex.setCone(7, [3, 4])
plex.setCone(8, [4, 2])
plex.setCone(9, [4, 5])
plex.setCone(10, [5, 3])
plex.symmetrize()
plex.stratify()
plex.view()
DM Object: 1 MPI process
type: plex
DM_0xc406d60_0 in 2 dimensions:
Number of 0-cells per rank: 4
Number of 1-cells per rank: 5
Number of 2-cells per rank: 2
Labels:
depth: 3 strata with value/size (0 (4), 1 (5), 2 (2))
celltype: 3 strata with value/size (0 (4), 1 (5), 3 (2))
9.1.2. 使用高级接口#
cells = [
[0, 1, 2],
[1, 3, 2]
]
coords = [
[-1, 0],
[0, -1],
[0, 1],
[1, 0]
]
plex = PETSc.DMPlex().createFromCellList(
dim=2, cells=cells, coords=coords, interpolate=True, comm=None)
plex.view()
DM Object: DM_0xc406d60_1 1 MPI process
type: plex
DM_0xc406d60_1 in 2 dimensions:
Number of 0-cells per rank: 4
Number of 1-cells per rank: 5
Number of 2-cells per rank: 2
Labels:
celltype: 3 strata with value/size (0 (4), 1 (5), 3 (2))
depth: 3 strata with value/size (0 (4), 1 (5), 2 (2))
9.2. PETSc.Section#
section1 = PETSc.Section().create()
section1.setChart(*plex.getChart())
section2 = PETSc.Section().create()
section2.setChart(*plex.getChart())
old_to_new = plex.getOrdering(PETSc.Mat.OrderingType.RCM).indices
reordering = np.empty_like(old_to_new) # reordering[new] -> old
reordering[old_to_new] = np.arange(old_to_new.size, dtype=old_to_new.dtype)
perm = PETSc.IS().createGeneral(reordering)
section2.setPermutation(perm)
ps, pe = plex.getDepthStratum(0)
for p in range(ps, pe):
section1.addDof(p, 1)
section2.addDof(p, 1)
section1.setUp()
section2.setUp()
section1.view()
section2.view()
PetscSection Object: 1 MPI process
type not yet set
Process 0:
( 0) dof 0 offset 0
( 1) dof 0 offset 0
( 2) dof 1 offset 0
( 3) dof 1 offset 1
( 4) dof 1 offset 2
( 5) dof 1 offset 3
( 6) dof 0 offset 4
( 7) dof 0 offset 4
( 8) dof 0 offset 4
( 9) dof 0 offset 4
( 10) dof 0 offset 4
PetscSection Object: 1 MPI process
type not yet set
Process 0:
( 0) dof 0 offset 0
( 1) dof 0 offset 0
( 2) dof 1 offset 3
( 3) dof 1 offset 0
( 4) dof 1 offset 2
( 5) dof 1 offset 1
( 6) dof 0 offset 4
( 7) dof 0 offset 4
( 8) dof 0 offset 4
( 9) dof 0 offset 4
( 10) dof 0 offset 4
9.3. Firedrake 中的 Mesh#
mesh = RectangleMesh(4, 4, 1, 1)
mesh.init()
plex = mesh.topology_dm
rank, size = mesh.comm.rank, mesh.comm.size
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[11], line 2
1 mesh = RectangleMesh(4, 4, 1, 1)
----> 2 mesh.init()
3 plex = mesh.topology_dm
4 rank, size = mesh.comm.rank, mesh.comm.size
File /opt/firedrake/firedrake/mesh.py:2811, in MeshGeometry.__getattr__(self, name)
2810 def __getattr__(self, name):
-> 2811 val = getattr(self._topology, name)
2812 setattr(self, name, val)
2813 return val
AttributeError: 'MeshTopology' object has no attribute 'init'
%%px --block
from firedrake import *
from firedrake.petsc import PETSc
from mpi4py import MPI
mesh = RectangleMesh(4, 4, 1, 1)
mesh.init()
plex = mesh.topology_dm
rank, size = mesh.comm.rank, mesh.comm.size
%%px --block
mesh._dm_renumbering.indices
和重新编号相关的函数
%%px --block
# https://petsc.org/release/manualpages/DMPlex/DMPlexGetOrdering
old_to_new = plex.getOrdering(PETSc.Mat.OrderingType.RCM).indices
reordering = np.empty_like(old_to_new)
reordering[old_to_new] = np.arange(old_to_new.size, dtype=old_to_new.dtype)
mesh._default_reordering, reordering
%%px --block
mesh.topology_dm.view()
entity_classes = mesh._entity_classes
ec_msg = ', '.join([f'{i}-cells: {ec}' for i, ec in enumerate(entity_classes)])
PETSc.Sys.syncPrint(f'[{rank}/{size}] mesh._entity_classes (core, owned, ghost): {ec_msg}')
PETSc.Sys.syncFlush()
计算 node_set 相关的函数
计算 node_set 相关的概念
V1 = FunctionSpace(mesh, 'CG', 1)
V2 = FunctionSpace(mesh, 'CG', 2)
PETSc.Sys.Print(f'V1 entity_dofs: {V1.finat_element.entity_dofs()}')
PETSc.Sys.Print(f'V2 entity_dofs: {V2.finat_element.entity_dofs()}')
nodes_per_entity_V1 = tuple(mesh.make_dofs_per_plex_entity(V1.finat_element.entity_dofs()))
nodes_per_entity_V2 = tuple(mesh.make_dofs_per_plex_entity(V2.finat_element.entity_dofs()))
PETSc.Sys.Print(f'V1 nodes_per_entity: {nodes_per_entity_V1}')
PETSc.Sys.Print(f'V2 nodes_per_entity: {nodes_per_entity_V2}')
%%px --block
V1 = FunctionSpace(mesh, 'CG', 1)
V2 = FunctionSpace(mesh, 'CG', 2)
PETSc.Sys.Print(f'V1 entity_dofs: {V1.finat_element.entity_dofs()}')
PETSc.Sys.Print(f'V2 entity_dofs: {V2.finat_element.entity_dofs()}')
nodes_per_entity_V1 = tuple(mesh.make_dofs_per_plex_entity(V1.finat_element.entity_dofs()))
nodes_per_entity_V2 = tuple(mesh.make_dofs_per_plex_entity(V2.finat_element.entity_dofs()))
PETSc.Sys.Print(f'V1 nodes_per_entity: {nodes_per_entity_V1}')
PETSc.Sys.Print(f'V2 nodes_per_entity: {nodes_per_entity_V2}')
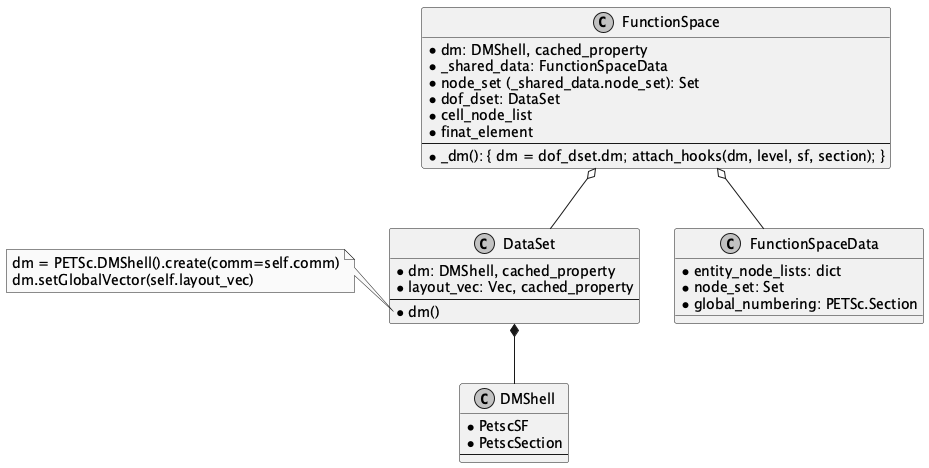
9.4. FunctionSpace#
下图使用 Sphinx / Jupyter Book 图形插件 plantuml 制作.
Fig. 9.1 class FunctionSpace#
9.5. 示例: 寻找特定的几何实体以及节点编号#
9.5.1. 二维示例#
使用 global_numbering 寻找某条边界上端点, 以及在这条边界上与它相邻的点 (2D mesh)
from firedrake import *
import matplotlib.pyplot as plt
import numpy as np
rectangle = Mesh("gmsh/rectangle.msh")
fig, axes = plt.subplots(figsize=[4, 3])
triplot(rectangle, axes=axes)
axes.set_aspect('equal')
rectangle.topology_dm.view()
%%px --block
from firedrake import *
import matplotlib.pyplot as plt
import numpy as np
rectangle = Mesh("gmsh/rectangle.msh")
fig, axes = plt.subplots(figsize=[4, 3])
triplot(rectangle, axes=axes)
axes.set_aspect('equal')
axes.set_xlim([-0.1, 1.1])
axes.set_ylim([-0.1, 1.1])
rectangle.topology_dm.view()
def get_interface_element_with_contact_point(mesh, V, interface_tag, adj_line_tag):
dm = mesh.topology_dm
edge_label = dm.getLabel("Face Sets") # 2D Face is Edge
core_label = dm.getLabel("pyop2_core")
owned_label = dm.getLabel("pyop2_owned")
edge_label_values = edge_label.getValueIS().indices
interface_indices = []
if interface_tag in edge_label_values:
interface_indices = edge_label.getStratumIS(interface_tag).indices
adj_line = []
if adj_line_tag in edge_label_values:
adj_line = edge_label.getStratumIS(adj_line_tag).indices
points = np.intersect1d(interface_indices, adj_line)
plex_element = []
if len(points) > 0:
point = points[0]
support = dm.getSupport(point)
for edge in support:
if edge_label.getValue(edge) == interface_tag and \
(core_label.getValue(edge) == 1 or owned_label.getValue(edge) == 1):
cone = dm.getCone(edge)
adj_point = cone[1] if cone[0] == point else cone[0]
plex_element = [point, adj_point]
break
local_section = V.global_numbering # global_numbering is a local section
element = [local_section.getOffset(_) for _ in plex_element]
return element
# interface_tag, adj_line_tag = 2, 4 # 2: left boundary, 4: upper boundary
V = FunctionSpace(rectangle, 'CG', 1)
element = get_interface_element_with_contact_point(rectangle, V, interface_tag=1, adj_line_tag=3)
coords_data = rectangle.coordinates.dat.data_ro_with_halos # This must be outside the if condition (mpi collective)
rank, size = rectangle.comm.rank, rectangle.comm.size
if len(element) > 0:
coords = [coords_data[_] for _ in element]
PETSc.Sys.syncPrint(f"[{rank}/{size}] node {element[0]}: {coords[0]}), node {element[1]}: {coords[1]}")
PETSc.Sys.syncFlush()
%%px --block
def get_interface_element_with_contact_point(mesh, V, interface_tag, adj_line_tag):
dm = mesh.topology_dm
edge_label = dm.getLabel("Face Sets") # 2D Face is Edge
core_label = dm.getLabel("pyop2_core")
owned_label = dm.getLabel("pyop2_owned")
edge_label_values = edge_label.getValueIS().indices
interface_indices = []
if interface_tag in edge_label_values:
interface_indices = edge_label.getStratumIS(interface_tag).indices
adj_line = []
if adj_line_tag in edge_label_values:
adj_line = edge_label.getStratumIS(adj_line_tag).indices
points = np.intersect1d(interface_indices, adj_line)
plex_element = []
if len(points) > 0:
point = points[0]
support = dm.getSupport(point)
for edge in support:
if edge_label.getValue(edge) == interface_tag and \
(core_label.getValue(edge) == 1 or owned_label.getValue(edge) == 1):
cone = dm.getCone(edge)
adj_point = cone[1] if cone[0] == point else cone[0]
plex_element = [point, adj_point]
break
local_section = V.global_numbering # global_numbering is a local section
element = [local_section.getOffset(_) for _ in plex_element]
return element
# interface_tag, adj_line_tag = 2, 4 # 2: left boundary, 4: upper boundary
V = FunctionSpace(rectangle, 'CG', 1)
element = get_interface_element_with_contact_point(rectangle, V, interface_tag=2, adj_line_tag=4)
coords_data = rectangle.coordinates.dat.data_ro_with_halos # This must be outside the if condition (mpi collective)
rank, size = rectangle.comm.rank, rectangle.comm.size
if len(element) > 0:
coords = [coords_data[_] for _ in element]
PETSc.Sys.syncPrint(f"[{rank}/{size}] node {element[0]}: {coords[0]}), node {element[1]}: {coords[1]}")
PETSc.Sys.syncFlush()
9.5.2. 三维示例#
使用 global_numbering 寻找界面上与接触线相邻的三角形 (3D mesh)
本示例网格文件 cylinder.msh 由几何文件 cylinder.geo 生成,
如图: 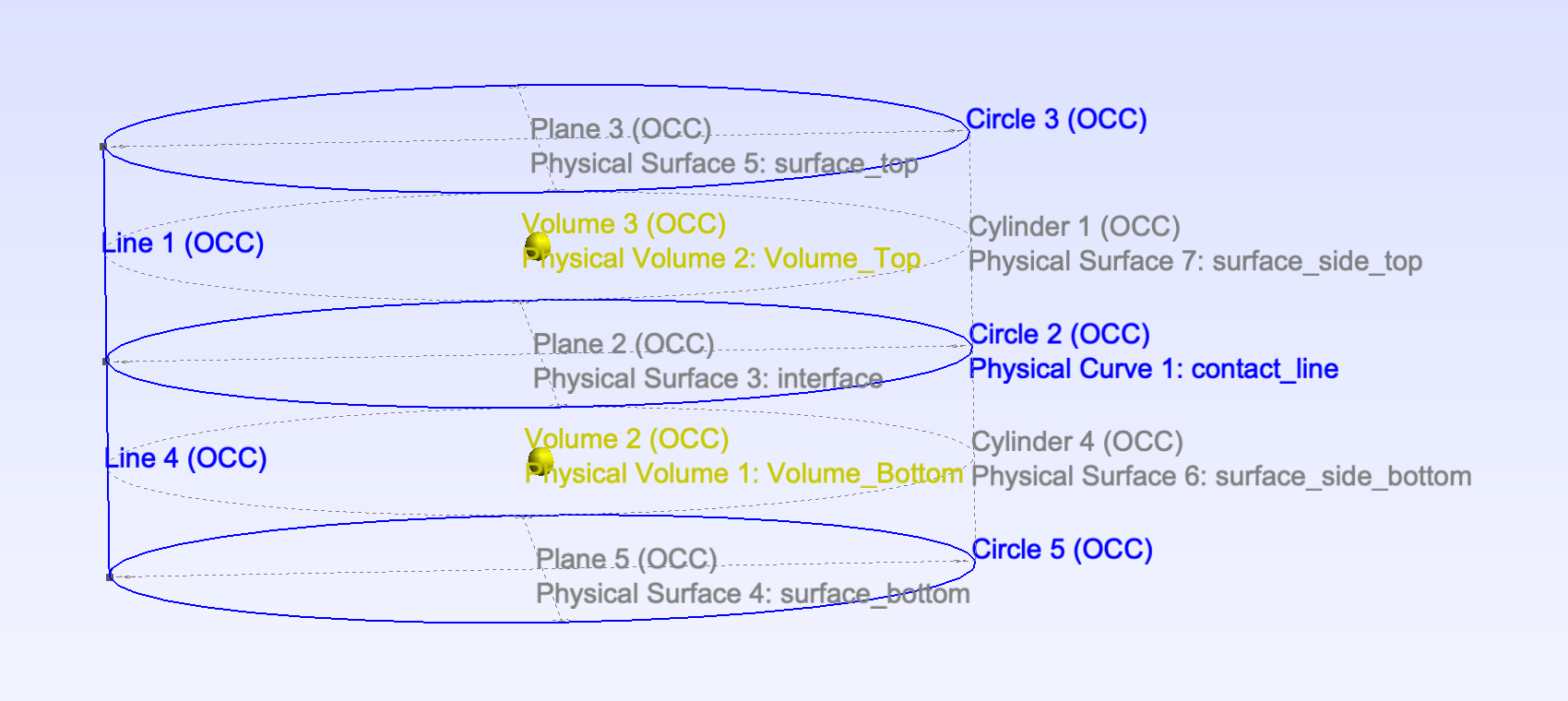
import matplotlib.pyplot as plt
import numpy as np
cylinder = Mesh("gmsh/cylinder.msh")
fig, axes = plt.subplots(figsize=[4, 3], subplot_kw={'projection': '3d'})
triplot(cylinder, axes=axes)
axes.set_aspect('equal')
cylinder.topology_dm.view()
%%px --block
import matplotlib.pyplot as plt
import numpy as np
cylinder = Mesh("gmsh/cylinder.msh")
fig, axes = plt.subplots(figsize=[4, 3], subplot_kw={'projection': '3d'})
triplot(cylinder, axes=axes)
axes.set_aspect('equal')
cylinder.topology_dm.view()
def get_interface_element_include_contact_line(mesh, V, interface_tag, contact_line_tag):
dm = mesh.topology_dm
edge_label = dm.getLabel("Edge Sets")
face_label = dm.getLabel("Face Sets")
core_label = dm.getLabel("pyop2_core")
owned_label = dm.getLabel("pyop2_owned")
edge_label_values = edge_label.getValueIS().indices
contact_line = []
if contact_line_tag in edge_label_values:
contact_line = edge_label.getStratumIS(contact_line_tag).indices
faces = []
for seg in contact_line:
for face in dm.getSupport(seg):
if face_label.getValue(face) == interface_tag and \
(core_label.getValue(face) == 1 or owned_label.getValue(face) == 1):
faces.append(int(face))
break
plex_cell_node_map = np.zeros((len(faces), 3), dtype=np.int32)
for i, (seg, face) in enumerate(zip(contact_line, faces)):
seg_nodes = dm.getCone(seg)
plex_cell_node_map[i, :2] = seg_nodes
plex_cell_node_map[i, 2:] = np.setdiff1d(
np.unique(np.array([dm.getCone(_) for _ in dm.getCone(face)]).flatten()),
seg_nodes)
local_section = V.global_numbering # global_numbering is a local section
cell_node_map = np.zeros_like(plex_cell_node_map)
for i, cell in enumerate(plex_cell_node_map):
assert np.all(np.array([local_section.getDof(_) for _ in cell]) > 0)
cell_node_map[i, :] = [local_section.getOffset(_) for _ in cell]
return cell_node_map
CONTACT_LINE = 1
INTERFACE = 3
V = FunctionSpace(cylinder, 'CG', 1)
cell_node_map = get_interface_element_include_contact_line(cylinder, V, INTERFACE, CONTACT_LINE)
%%px --block
def get_interface_element_include_contact_line(mesh, V, interface_tag, contact_line_tag):
dm = mesh.topology_dm
edge_label = dm.getLabel("Edge Sets")
face_label = dm.getLabel("Face Sets")
core_label = dm.getLabel("pyop2_core")
owned_label = dm.getLabel("pyop2_owned")
edge_label_values = edge_label.getValueIS().indices
contact_line = []
if contact_line_tag in edge_label_values:
contact_line = edge_label.getStratumIS(contact_line_tag).indices
faces = []
for seg in contact_line:
for face in dm.getSupport(seg):
if face_label.getValue(face) == interface_tag and \
(core_label.getValue(face) == 1 or owned_label.getValue(face) == 1):
faces.append(int(face))
break
plex_cell_node_map = np.zeros((len(faces), 3), dtype=np.int32)
for i, (seg, face) in enumerate(zip(contact_line, faces)):
seg_nodes = dm.getCone(seg)
plex_cell_node_map[i, :2] = seg_nodes # set the seg nodes first
plex_cell_node_map[i, 2:] = np.setdiff1d(
np.unique(np.array([dm.getCone(_) for _ in dm.getCone(face)]).flatten()),
seg_nodes)
local_section = V.global_numbering # global_numbering is a local section
cell_node_map = np.zeros_like(plex_cell_node_map)
for i, cell in enumerate(plex_cell_node_map):
assert np.all(np.array([local_section.getDof(_) for _ in cell]) > 0)
cell_node_map[i, :] = [local_section.getOffset(_) for _ in cell]
return cell_node_map
CONTACT_LINE = 1
INTERFACE = 3
V = FunctionSpace(cylinder, 'CG', 1)
cell_node_map = get_interface_element_include_contact_line(cylinder, V, INTERFACE, CONTACT_LINE)
# plot the triangle to check if they are on the interface
coords = cylinder.coordinates.dat.data_ro_with_halos
if len(cell_node_map) > 0:
assert np.allclose(coords[:, 2][cell_node_map], 0.25)
fig, axes = plt.subplots(figsize=[4, 3])
c = axes.triplot(coords[:, 0], coords[:, 1], triangles=cell_node_map)
lines = [[(coords[_, 0], coords[_, 1]) for _ in __[:2] ] for __ in cell_node_map]
from matplotlib.collections import LineCollection
line_collection = LineCollection(lines, colors='k', linestyles=':')
axes.add_collection(line_collection)
axes.set_xlim([-0.52, 0.52])
axes.set_ylim([-0.52, 0.52])
axes.set_aspect("equal")
axes.grid("on")
%%px --block
# plot the triangle to check if they are on the interface
coords = cylinder.coordinates.dat.data_ro_with_halos
if len(cell_node_map) > 0:
assert np.allclose(coords[:, 2][cell_node_map], 0.25)
fig, axes = plt.subplots(figsize=[4, 3])
c = axes.triplot(coords[:, 0], coords[:, 1], triangles=cell_node_map)
lines = [[(coords[_, 0], coords[_, 1]) for _ in __[:2] ] for __ in cell_node_map]
from matplotlib.collections import LineCollection
line_collection = LineCollection(lines, colors='k', linestyles=':')
axes.add_collection(line_collection)
axes.set_xlim([-0.52, 0.52])
axes.set_ylim([-0.52, 0.52])
axes.set_aspect("equal")
axes.grid("on")