7. PETSc#
PETSc, the Portable, Extensible Toolkit for Scientific Computation, pronounced PET-see (/ˈpɛt-siː/), is for the scalable (parallel) solution of scientific applications modeled by partial differential equations.
PETSc 是由阿贡国家实验室开发的便携可扩展科学计算工具包, 提供了并行求解大规模方程组的许多算法, 并且可调用外部包对方程组进行求解. 另外, PETSc 也提供了用于数值求解偏微分方程的组件, 包括结构化网格数据结构, 非结构化网格数据结构, 和有限元空间等.
Firedrake 可以看作是 PETSc 的高层次封装. 在 Firedrake 中, 一般无需操作 PETSc 对象, 但有些特殊情况必须直接操纵 PETSc 对象, 并且有时候直接操纵 PETSc 对象会更高效.
PETSc 是 c 语言包, 也提供了 Fortran 接口. petsc4py 是 PETSc 的 python 封装.
学习 PETSc 可以从 Texas Advanced Computing Center (TACC) 发布的 PETSc 入门课程开始
PETSc 网站的手册和入门讲义
PETSc 代码库有许多示例可以作为学习材料, 如 PETSc 仓库中 petsc4py 的示例:
Tip
PETSc 目录中有用的工具, 如 h5dump, petsc_gen_xdmf.py, PetscBinaryIO.py 等.
在 PETSc 环境中, 运行如下命令添加这些工具所在路径到 PATH:
export PATH="$PATH:$PETSC_DIR/lib/petsc/bin"
export PATH="$PATH:$PETSC_DIR/${PETSC_ARCH-default}/bin"
在激活的 Firedrake 环境可以运行如下命令的输出, 添加工具所在路径到环境变量 PATH.
python -c "from firedrake import *; \
import os; \
PETSC_DIR = os.environ['PETSC_DIR']; \
PETSC_ARCH = os.environ['PETSC_ARCH']; \
print('\nRun the follwoing code to add petsc/bin to path:\n'); \
print(f' export PATH=\"\$PATH:{PETSC_DIR}/lib/petsc/bin\"'); \
print(f' export PATH=\"\$PATH:{PETSC_DIR}/{PETSC_ARCH}/bin\"'); \
print('');"
7.1. Vector and Matirx#
保存矩阵到文件: matvecio.py
from firedrake import *
from firedrake.petsc import PETSc
test_mesh = RectangleMesh(nx=4, ny=4, Lx=1, Ly=1)
x, y = SpatialCoordinate(test_mesh)
f = sin(pi*x)*sin(pi*y)
V = FunctionSpace(test_mesh, 'CG', degree=1)
u, v = TrialFunction(V), TestFunction(V)
a = inner(grad(u), grad(v))*dx
L = inner(f, v)*dx
A = assemble(a)
b = assemble(L)
type(A), type(b)
(firedrake.matrix.Matrix, firedrake.function.Function)
7.1.1. Matrix#
type(A.petscmat)
petsc4py.PETSc.Mat
单进程运行且矩阵不大时, 可以把 PETSc 矩阵转换为 numpy 数组
import numpy as np
from scipy.sparse import csr_matrix
m, n = A.petscmat.getSize()
indptr, indices, data = A.petscmat.getValuesCSR()
A_numpy = csr_matrix((data, indices, indptr), shape=(m, n)).toarray()
A.petscmat.getRow(0), A_numpy[0, :]
((array([0, 1, 2], dtype=int32), array([ 1. , -0.5, -0.5])),
array([ 1. , -0.5, -0.5, 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. ]))
保存矩阵到文件 MatViewFromOptions
https://petsc.org/main/manualpages/Mat/MatViewFromOptions/
在代码中加入如下行
A.petscmat.viewFromOptions('-A_view')
那么在命令行可以通过选项 -A_view binary:A.bin 保存 A 到文件 A.bin.
7.1.2. Convert petsc matrix to csr_matrix and compute determinant#
# %load py/poisson_example1.py
from firedrake import *
from firedrake.petsc import PETSc
from scipy.sparse import csr_matrix
from scipy.sparse.linalg import splu
import numpy as np
N = 8
test_mesh = RectangleMesh(nx=N, ny=N, Lx=1, Ly=1)
x, y = SpatialCoordinate(test_mesh)
f = sin(pi*x)*sin(pi*y)
g = Constant(0)
V = FunctionSpace(test_mesh, 'CG', degree=1)
u, v = TrialFunction(V), TestFunction(V)
a = inner(grad(u), grad(v))*dx
L = inner(f, v)*dx # or f*v*dx
bc = DirichletBC(V, g=g, sub_domain='on_boundary')
u_h = Function(V, name='u_h')
problem = LinearVariationalProblem(a, L, u_h, bcs=bc)
solver = LinearVariationalSolver(problem, options_prefix='')
solver.solve()
ksp = solver.snes.getKSP()
A, P = ksp.getOperators()
m, n = A.getSize()
indptr, indices, data = A.getValuesCSR()
A_numpy = csr_matrix((data, indices, indptr), shape=(m, n)).toarray()
print(A_numpy)
print(data)
lu = splu(A_numpy)
diagL = lu.L.diagonal()
diagU = lu.U.diagonal()
det=diagL.prod()*diagU.prod()
print(det)
mpiexec --bind-to none -n 1 \
python3 a.py \
-ksp_view \
-ksp_error_if_not_converged \
-mat_mumps_icntl_33 1 \
-mat_mumps_icntl_4 4 \
-ksp_monitor 2>&1 | tee "my-$(date +%Y%m%d-%H:%M:%S).log"
7.1.3. Vector#
with b.dat.vec_ro as vec:
print(type(vec))
<class 'petsc4py.PETSc.Vec'>
7.1.4. ISLocalToGlobalMapping#
Create a local to global map
import firedrake as fd
from firedrake.petsc import PETSc
from pyop2.datatypes import IntType, ScalarType
import numpy as np
rank = COMM_WORLD.rank
owned_sz = np.array(rank+3, dtype=IntType)
offset = np.empty_like(owned_sz)
COMM_WORLD.Scan(owned_sz, offset)
offset -= owned_sz
indices = np.arange(offset, offset + owned_sz, dtype=IntType)
lgmap = PETSc.LGMap()
lgmap.create(indices, bsize=1, comm=COMM_WORLD)
lgmap.view()
ISLocalToGlobalMapping Object: 1 MPI process
type not yet set
[0] 0 0
[0] 1 1
[0] 2 2
7.2. KSP#
自定义 KSP 进行线性方程组求解请参考 PETSc 的文档
求解完成需要检查是否收敛
from firedrake.exceptions import ConvergenceError
from firedrake.petsc import OptionsManager, PETSc
from firedrake.solving_utils import KSPReasons
import numpy as np
def _make_reasons(reasons):
return dict([(getattr(reasons, r), r)
for r in dir(reasons) if not r.startswith('_')])
PCFailedReason = _make_reasons(PETSc.PC.FailedReason())
def get_ksp_reason(ksp):
r = ksp.getConvergedReason()
pc = ksp.getPC()
r_pc = pc.getFailedReason()
return KSPReasons[r], PCFailedReason[r_pc]
A = PETSc.Mat()
A.create(PETSc.COMM_WORLD)
A.setSizes([2, 2])
A.setType('aij') # sparse
# A.setPreallocationNNZ(4)
A.setUp()
A.setValue(1, 0, 1)
A.setValue(0, 1, np.inf) # to make the solver failed
A.assemble()
ksp = PETSc.KSP().create()
ksp.setOperators(A) # solve A*x=b by ksp.solve(b,x)
om = OptionsManager(
{
'ksp_type': 'preonly',
'pc_type': 'lu',
# 'ksp_view': None,
'pc_factor_mat_solver_type': 'mumps',
# 'ksp_error_if_not_converged': None,
},
options_prefix='test')
om.set_from_options(ksp)
x, b = A.createVecs()
b.setValue(0, 1)
# ksp.view()
with om.inserted_options():
try:
ksp.solve(b, x)
r = ksp.getConvergedReason()
if r < 0:
raise ConvergenceError(KSPReasons[r])
except ConvergenceError as e:
r, r_pc = get_ksp_reason(ksp)
PETSc.Sys.Print(f"Error: solver did not converged: {r}, PC: {r_pc}")
except PETSc.Error as e:
if e.ierr == 91: # https://petsc.org/release/include/petscerror.h.html
PETSc.Sys.Print(f"Error from PETSc: solver did not converged: {KSPReasons[ksp.getConvergedReason()]}")
elif e.ierr == 76:
PETSc.Sys.Print(f"Error from PETSc:")
PETSc.Sys.Print(f" ksp reason: {KSPReasons[ksp.getConvergedReason()]}")
PETSc.Sys.Print(f" error in library called by PETSc:")
PETSc.Sys.Print(" "*4 + str(e).replace("\n", "\n" + " "*4))
# We should terminate the process when an error occured in petsc
# as suggested by Matt https://lists.mcs.anl.gov/pipermail/petsc-users/2023-March/048146.html
raise
Error: solver did not converged: DIVERGED_PCSETUP_FAILED, PC: FACTOR_NUMERIC_ZEROPIVOT
查看特征值和残差变化, 并保存图片
python test.py -ksp_type gmres -pc_type jacobi -ksp_view_eigenvalues draw -ksp_monitor draw::draw_lg -draw_save .png
7.2.1. Check ksp status in Firedrake#
from firedrake import *
from firedrake.petsc import PETSc
from firedrake.solving_utils import KSPReasons
import numpy as np
def printf(*args, **kwargs):
PETSc.Sys.Print(*args, **kwargs)
def get_ksp_reason(solver):
r = solver.snes.getKSP().getConvergedReason()
return KSPReasons[r]
rank, size = COMM_WORLD.rank, COMM_WORLD.size
opts = PETSc.Options()
N = opts.getInt('N', 32*size)
test_mesh = RectangleMesh(nx=N, ny=N, Lx=1, Ly=1)
x, y = SpatialCoordinate(test_mesh)
f = sin(pi*x)*sin(pi*y)
V = FunctionSpace(test_mesh, 'CG', degree=1)
u, v = TrialFunction(V), TestFunction(V)
a = inner(grad(u), grad(v))*dx - inner(f, v)*dx
bc = DirichletBC(V, 0, sub_domain='on_boundary')
u_h = Function(V, name='u_h')
problem = LinearVariationalProblem(lhs(a), rhs(a), u_h, bcs=bc)
solver_parameters = {'ksp_type': 'cg',
'ksp_max_it': 4,
'ksp_converged_reason': None,
# 'ksp_error_if_not_converged': None,
'pc_type': 'none'}
solver = LinearVariationalSolver(problem, solver_parameters=solver_parameters, options_prefix='')
for i in range(3):
printf(f"Loop i = {i}")
try:
solver.solve()
except ConvergenceError:
printf(f" Error from Firedrake: solver did not converged: {get_ksp_reason(solver)}")
except PETSc.Error as e:
if e.ierr == 91: # https://petsc.org/release/include/petscerror.h.html
printf(f" Error from PETSc: solver did not converged: {get_ksp_reason(solver)}")
elif e.ierr == 76:
PETSc.Sys.Print(f"Error from PETSc:")
PETSc.Sys.Print(f" ksp reason: {KSPReasons[ksp.getConvergedReason()]}")
PETSc.Sys.Print(f" error in library called by PETSc:")
PETSc.Sys.Print(" "*4 + str(e).replace("\n", "\n" + " "*4))
# We should terminate the process when an error occured in petsc
# as suggested by Matt https://lists.mcs.anl.gov/pipermail/petsc-users/2023-March/048146.html
raise
Loop i = 0
Linear solve did not converge due to DIVERGED_ITS iterations 4
Error from Firedrake: solver did not converged: DIVERGED_MAX_IT
Loop i = 1
Linear solve did not converge due to DIVERGED_ITS iterations 4
Error from Firedrake: solver did not converged: DIVERGED_MAX_IT
Loop i = 2
Linear solve did not converge due to DIVERGED_ITS iterations 4
Error from Firedrake: solver did not converged: DIVERGED_MAX_IT
7.3. DMPlex#
DMPlex 是管理非结构化网格的数据结构, 内部使用有向无环图表示网格的拓扑关系 [HKA+21, LMKG16].
DMPlex 中的 Plex 表示 Complex. 并行计算时, 网格会被划分成不同的块, 分配到各个进程.
7.3.1. 网格粗化#
import sys
import petsc4py
petsc4py.init(sys.argv)
from petsc4py import PETSc
def output_vtk(dmplex, filename):
viewer = PETSc.Viewer().createVTK(filename, 'w')
viewer.view(dmplex)
opts = PETSc.Options()
N = opts.getInt('N', 4)
dim = opts.getInt('dim', 3)
overlap = opts.getInt('overlap', 1)
faces = [N for _ in range(dim)]
plex = PETSc.DMPlex().createBoxMesh(faces, simplex=True)
plex.setName('Init DM')
plex.viewFromOptions('-init_dm_view')
sf = plex.distribute(overlap=overlap)
plex.setName('Distribue DM')
plex.viewFromOptions('-dist_dm_view')
new_plex = plex.coarsen()
new_plex.setName('Coarsen DM')
new_plex.viewFromOptions('-coarsen_dm_view')
# mpiexec -n 2 python test_coarsen.py -dim 3 -overlap 0 -dm_adaptor parmmg -coarsen_dm_view vtk:data/test.vtu
7.4. Viewer#
Option values for Viewer:
If no value is provided ascii:stdout is used
ascii[:[filename][:[format][:append]]] defaults to stdout - format can be one of ascii_info, ascii_info_detail, or ascii_matlab,
for example ascii::ascii_info prints just the information about the object not all details
unless :append is given filename opens in write mode, overwriting what was already there
binary[:[filename][:[format][:append]]] defaults to the file binaryoutput
draw[:drawtype[:filename]] for example, draw:tikz, draw:tikz:figure.tex or draw:x
socket[:port] defaults to the standard output port
saws[:communicatorname] publishes object to the Scientific Application
Webserver (SAWs)
-nox - do not use X graphics (ignore graphics calls, but run program correctly)
-nox_warning - when X Windows support is not installed this prevents the warning message from being printed
-draw_pause <pause amount> -- -1 indicates wait for mouse input,
-2 indicates pause when window is to be destroyed
-draw_marker_type - <x,point>
-draw_save [optional filename] - (X Windows only) saves each image before it is cleared to a file
-draw_save_final_image [optional filename] - (X Windows only) saves the final image displayed in a window
-draw_save_movie - converts image files to a movie at the end of the run. See PetscDrawSetSave()
-draw_save_single_file - saves each new image in the same file, normally each new image is saved in a new file with 'filename/filename_%d.ext'
-draw_save_on_clear - saves an image on each clear, mainly for debugging
-draw_save_on_flush - saves an image on each flush, mainly for debugging
7.4.1. Load mesh file and view (petsc4py)#
import sys
import petsc4py
petsc4py.init(sys.argv)
from petsc4py import PETSc
import numpy as np
dm = PETSc.DMPlex().createFromFile('gmsh/Lshape.msh', plexname='test')
dm.view()
# hdf5 for load
viewer = PETSc.Viewer().createHDF5('data/Lshape.h5', mode='w')
viewer(dm)
# hdf5 for visualization:
# You can generate xdmf file from this file by
# `petsc/lib/petsc/bin/petsc_gen_xdmf.py`
# Then load the xdmf file to paraview to visualize the mesh.
viewer = PETSc.Viewer().createHDF5('data/Lshape_xdmf.h5', mode='w')
viewer.pushFormat(viewer.Format.HDF5_XDMF)
viewer(dm)
viewer.popFormat()
# vtk file
viewer = PETSc.Viewer().createVTK('data/Lshape.vtk', mode='w')
viewer(dm)
# draw on X window
# viewer = PETSc.Viewer().createDraw()
# viewer(dm)
DM Object: test 1 MPI process
type: plex
test in 2 dimensions:
Number of 0-cells per rank: 274
Number of 1-cells per rank: 755
Number of 2-cells per rank: 482
Labels:
celltype: 3 strata with value/size (0 (274), 3 (482), 1 (755))
depth: 3 strata with value/size (0 (274), 1 (755), 2 (482))
# since the petsc_draw is not in petsc4py, we use options to save the images
opts = PETSc.Options()
opts_old = opts.getAll()
opts.insertString('-dm_view draw:tikz:data/Lshape.tex')
dm.viewFromOptions('-dm_view')
7.4.2. View mesh of firedrake by DMPlex#
%%px --block
from firedrake import *
mesh = RectangleMesh(8, 8, 1, 1)
mesh.topology_dm.view()
[stdout:0] DM Object: firedrake_default_topology 2 MPI processes
type: plex
firedrake_default_topology in 2 dimensions:
Number of 0-cells per rank: 45 45
Number of 1-cells per rank: 108 108
Number of 2-cells per rank: 64 64
Labels:
depth: 3 strata with value/size (0 (45), 1 (108), 2 (64))
celltype: 3 strata with value/size (0 (45), 1 (108), 3 (64))
Face Sets: 2 strata with value/size (1 (8), 3 (8))
exterior_facets: 1 strata with value/size (1 (16))
interior_facets: 1 strata with value/size (1 (92))
7.5. Star Forest 结构体 PetscSF#
PetscSF 是 PETSc 中用于进程间数据交换的数据结构.
它存储了根节点(当前进程上)和叶子节点(任意进程上)的对应关系, 可以把数据从根节点发送到叶子节点, 也可以把数据从叶子节点收集到根节点 [ZBB+22].
%%px --block
from firedrake import *
from firedrake.petsc import PETSc
from petsc4py import PETSc
import numpy as np
# 6--------7--------8
# | | |
# 3--------4--------5
# | | |
# 0--------1--------2
def test_SFDistributeSection():
comm = COMM_WORLD
if comm.rank == 0:
cells = np.asarray(
[[0, 1, 3],
[1, 2, 4],
[1, 4, 3],
[2, 5, 4],
[3, 4, 6],
[4, 5, 7],
[4, 7, 6],
[5, 8, 7]], dtype=np.int32)
coords = np.asarray(
[[0. , 0. ],
[0.5, 0. ],
[1. , 0. ],
[0. , 0.5],
[0.5, 0.5],
[1.0, 0.5],
[0. , 1. ],
[0.5, 1. ],
[1. , 1. ]], dtype=np.double)
else:
cells = np.zeros([0, 3], dtype=np.int32)
coords = np.zeros([0, 2], dtype=np.double)
dim = 2
plex = PETSc.DMPlex().createFromCellList(dim, cells, coords, comm=comm)
rootSection = PETSc.Section().create(comm=comm)
pStart, pEnd = plex.getHeightStratum(2)
rootSection.setChart(*plex.getChart())
for p in range(pStart, pEnd):
rootSection.setDof(p, 1)
rootSection.setUp()
rootSection.viewFromOptions('-section_view')
dplex = plex.clone()
msf = dplex.distribute()
if msf is None:
PETSc.Sys.Print("Warning: plex has not been distributed!")
return
dplex.viewFromOptions('-dm_view')
def isEqualSF(ssf0, ssf1):
nroots0, local0, remote0 = ssf0.getGraph()
nroots1, local1, remote1 = ssf1.getGraph()
return (nroots0 == nroots1) \
and np.array_equal(local0, local1) \
and np.array_equal(remote0, remote1)
remoteOffsets0, leafSection0 = msf.distributeSection(rootSection)
ssf0 = msf.createSectionSF(rootSection, remoteOffsets0, leafSection0)
remoteOffsets1, leafSection1 = msf.distributeSection(rootSection, None)
ssf1 = msf.createSectionSF(rootSection, remoteOffsets1, leafSection1)
leafSection2 = PETSc.Section()
remoteOffsets2, leafSection2 = msf.distributeSection(rootSection, leafSection2)
ssf2 = msf.createSectionSF(rootSection, remoteOffsets2, leafSection2)
leafSection3 = PETSc.Section()
remoteOffsets3, _ = msf.distributeSection(rootSection, leafSection3)
ssf3 = msf.createSectionSF(rootSection, remoteOffsets3, leafSection3)
leafSection4 = PETSc.Section().create(dplex.getComm())
remoteOffsets4, leafSection4 = msf.distributeSection(rootSection, leafSection4)
ssf4 = msf.createSectionSF(rootSection, remoteOffsets4, leafSection4)
leafSection5 = PETSc.Section().create(dplex.getComm())
remoteOffsets5, _ = msf.distributeSection(rootSection, leafSection5)
ssf5 = msf.createSectionSF(rootSection, remoteOffsets5, leafSection5)
assert isEqualSF(ssf0, ssf1)
assert isEqualSF(ssf0, ssf2)
assert isEqualSF(ssf0, ssf3)
assert isEqualSF(ssf0, ssf4)
ssf0.view()
%%px --block
# Add back after upgrad the firedrake
test_SFDistributeSection()
[stdout:0] PetscSF Object: 2 MPI processes
type: basic
[0] Number of roots=9, leaves=6, remote ranks=1
[0] 0 <- (0,0)
[0] 1 <- (0,1)
[0] 2 <- (0,3)
[0] 3 <- (0,4)
[0] 4 <- (0,6)
[0] 5 <- (0,7)
[1] Number of roots=0, leaves=6, remote ranks=1
[1] 0 <- (0,1)
[1] 1 <- (0,2)
[1] 2 <- (0,4)
[1] 3 <- (0,5)
[1] 4 <- (0,7)
[1] 5 <- (0,8)
MultiSF sort=rank-order
7.6. PCPatch#
PCPatch 在单元片上生成的预处理子. 用户可以使用 PETSc 提供的单元片类型 (star, vanka, pardecomp), 也可以自行构造.
它根据用户输入的关于解向量的 dm 和 sf 信息, 构造各个单元片上的自由度和全局自由度的对应关系. PCPatch 内部有以下变量
/* Topology */
PetscInt dim, codim; /* Dimension or codimension of mesh points to loop over; only one of them can be set */
PetscSection cellCounts; /* Maps patch -> # cells in patch */
IS cells; /* [patch][cell in patch]: Cell number */
PetscSection pointCounts; /* Maps patch -> # points with dofs in patch */
IS points; /* [patch][point in patch]: Point number */
PetscSection intFacetCounts;
PetscSection extFacetCounts;
PetscSection cellNumbering; /* Plex: NULL Firedrake: Numbering of cells in DM */
/* Dof layout */
PetscInt nsubspaces; /* Number of fields */
PetscSF sectionSF; /* Combined SF mapping process local to global */
PetscSection *dofSection; /* ?? For each field, patch -> # dofs in patch */
PetscInt *subspaceOffsets; /* Plex: NULL Firedrake: offset of each field in concatenated process local numbering for mixed spaces */
PetscInt **cellNodeMap; /* [field][cell][dof in cell]: global dofs in cell TODO Free this after its use in PCPatchCreateCellPatchDiscretisationInfo() */
IS dofs; /* [patch][cell in patch][dof in cell]: patch local dof */
IS offs; /* [patch][point in patch]: patch local offset (same layout as 'points', used for filling up patchSection) */
PetscSection gtolCounts; /* ?? Indices to extract from local to patch vectors */
IS gtol;
gtol 是从本进程自由度编号 (相对整个解来说是局部自由度, 但相对 patch 为全局自由度) 到单个 patch 上局部自由度的映射.
dofs Patch 上局部的自由度映射关系 (可以看作 patch 上的 cell_node_map, 和传入的 map 一致)
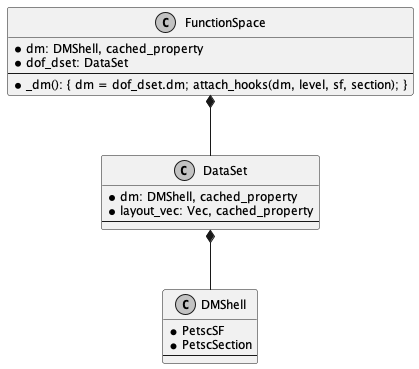
7.7. FunctionSpace in Firedrake#
Fig. 7.1 class FunctionSpace#
References
Vaclav Hapla, Matthew G. Knepley, Michael Afanasiev, Christian Boehm, Martin van Driel, Lion Krischer, and Andreas Fichtner. Fully parallel mesh i/o using petsc dmplex with an application to waveform modeling. SIAM Journal on Scientific Computing, 43(2):C127–C153, jan 2021. doi:10.1137/20m1332748.
Michael Lange, Lawrence Mitchell, Matthew G. Knepley, and Gerard J. Gorman. Efficient mesh management in firedrake using petsc dmplex. SIAM Journal on Scientific Computing, 38(5):S143–S155, jan 2016. doi:10.1137/15m1026092.
Junchao Zhang, Jed Brown, Satish Balay, Jacob Faibussowitsch, Matthew Knepley, Oana Marin, Richard Tran Mills, Todd Munson, Barry F. Smith, and Stefano Zampini. The PetscSF scalable communication layer. IEEE Transactions on Parallel and Distributed Systems, 33(4):842–853, apr 2022. doi:10.1109/tpds.2021.3084070.